name <- c("Can","Cem","Hande","Mehmet","Deniz","Kemal","Derya","Fatma")
gender <- c("Male","Male","Female","Male","Female","Male","Female","Female")
mode(gender)[1] "character"Consider the following data table:
| Name | Gender | Month of Birth |
|---|---|---|
| Can | Male | January |
| Cem | Male | July |
| Hande | Female | May |
| Mehmet | Male | May |
| Deniz | Female | February |
| Kemal | Male | July |
| Derya | Female | May |
| Fatma | Female | April |
All values in this data are strings. However, Gender and Month of Birth take only a limited set of values. They are categorical variables.
A categorical variable (or, a factor) can take one of predetermined, discrete values. Day of the week, month of the year, shirt size are categorical variables.
A single value of a categorical variable is called a level, such as “Monday”, “December”, or “XL”
First, let’s generate vectors to hold the relevant data.
name <- c("Can","Cem","Hande","Mehmet","Deniz","Kemal","Derya","Fatma")
gender <- c("Male","Male","Female","Male","Female","Male","Female","Female")
mode(gender)[1] "character"These vectors are of “character” type. We can convert the gender vector to a factor variable using the factor() function.
gender_fac <- factor(gender)
gender_fac[1] Male Male Female Male Female Male Female Female
Levels: Female Malemode(gender_fac)[1] "numeric"The mode() call on the factor variable returns numeric, because internally the levels are stored as integers 1,2,3… This fact makes it possible to add, remove, or rename levels.
The factor vector has an additional attribute, the levels information.
Get the vector of levels:
levels(gender_fac)[1] "Female" "Male" Get the number of levels:
nlevels(gender_fac)[1] 2Common R functions handle factors in specialized ways:
summary(gender) # character vector Length Class Mode
8 character character summary(gender_fac) # factorFemale Male
4 4 One can change the level names easily using an assignment to the levels() function.
levels(gender_fac) <- c("F","M")
gender_fac[1] M M F M F M F F
Levels: F MElements of factor-valued vectors are indexed in the same way as any other vector.
gender_fac[2:5][1] M F M F
Levels: F Mgender_fac[c(3,5,7:8)][1] F F F F
Levels: F MNote that the last result is composed of only F levels. However, the factor object still stores the the full set of levels F and M.
Our factor-valued vector:
gender_fac[1] M M F M F M F F
Levels: F MAs usual, the equality check returns a Boolean vector:
gender_fac=="M"[1] TRUE TRUE FALSE TRUE FALSE TRUE FALSE FALSEAmong the name vector, select elements with corresponding gender "M".
name[gender_fac=="M"][1] "Can" "Cem" "Mehmet" "Kemal" Sometimes we may want to remove one level in a category.
As an example, consider the following factor, where the level "Female" is misspelled twice as "female":
gender_fac <- factor(c("Male","Male","Female","Male","female","Male","female","Female"))
gender_fac[1] Male Male Female Male female Male female Female
Levels: female Female MaleThe resulting factor has three levels, but actually "female" and "Female" should be the same the same. Let’s fix this by overwriting all occurrences of "female" with "Female".
gender_fac[gender_fac=="female"] <- "Female"
gender_fac[1] Male Male Female Male Female Male Female Female
Levels: female Female MaleHowever, the levels attribute still lists the invalid "female" category. To remove it, we use the droplevels() function. It removes all levels for which there are no entries.
gender_fac <- droplevels(gender_fac)
gender_fac[1] Male Male Female Male Female Male Female Female
Levels: Female MaleConsider the following table again:
| Name | Gender | Month of Birth |
|---|---|---|
| Can | Male | January |
| Cem | Male | July |
| Hande | Female | May |
| Mehmet | Male | May |
| Deniz | Female | February |
| Kemal | Male | July |
| Derya | Female | May |
| Fatma | Female | April |
gender is an example of a nominal factor: There is no inherent order between levels. We cannot say whether “Male” is greater than “Female” or not.
On the other hand, month of birth information is an ordinal factor: Months have an inherent order, so it makes sense to say that “January” < “February”.
Suppose we use a vector to store the observed month-of-birth (MOB) data.
mob <- c("January","July","May","May","February","July","May","April")There are two problems with this vector:
mob[1] < mob[5][1] FALSEbecause the less-than operator compares with alphabetical order only.
Defining the data as a factor object solves both problems, if we set the levels properly. This can be done as follows:
months <- c("January","February","March","April","May",
"June","July","August","September","October","November","December")
mob_fac <- factor(mob, levels=months, ordered=TRUE)
mob_fac[1] January July May May February July May April
12 Levels: January < February < March < April < May < June < ... < DecemberThe ordered=TRUE setting ensures that this is an ordinal factor, and comparisons can be made in the same order levels are given.
mob_fac[1] < mob_fac[5] # January < February[1] TRUEThe summary() function gives a count of elements in each level.
summary(mob_fac) January February March April May June July August
1 1 0 1 3 0 2 0
September October November December
0 0 0 0 Earlier we have seen that combining two vectors into a single vector is done with the c() function:
x1 <- c(1,2,3,4)
x2 <- c(7,8,9)
c(x1, x2)[1] 1 2 3 4 7 8 9The same works with factor objects:
mob_fac[1] January July May May February July May April
12 Levels: January < February < March < April < May < June < ... < Decembermob2 <- factor(c("April","March","May"), levels=months, ordered=TRUE)
mob2[1] April March May
12 Levels: January < February < March < April < May < June < ... < Decemberc(mob_fac, mob2) [1] January July May May February July May April
[9] April March May
12 Levels: January < February < March < April < May < June < ... < DecemberOften, a continuous data is converted to a categorical variable. For example, age is converted to young/middle-aged/old, body size to small/medium/large, income to low/high.
Suppose we are given the following, numerical data:
x <- c(11, 18, 36, 74, 43, 81, 95, 64, 32, 51)We want to categorize this data as small for values in [0, 30), medium for [30, 70), and high for [70, 100].
The notation [30,70) means that the value 30 belongs to this category, but 70 does not.
The cut() function generates a factor object from continuous values. The breaks parameter is used to specify the intervals.
cut(x, breaks=c(0, 30, 70, 100)) [1] (0,30] (0,30] (30,70] (70,100] (30,70] (70,100] (70,100] (30,70]
[9] (30,70] (30,70]
Levels: (0,30] (30,70] (70,100]However, note that the ends of the intervals are not as we want. The first value of the boundary in not included in the interval, but the second value is.
To fix this, we set the parameter right to FALSE.
cut(x, breaks=c(0, 30, 70, 100), right = F) [1] [0,30) [0,30) [30,70) [70,100) [30,70) [70,100) [70,100) [30,70)
[9] [30,70) [30,70)
Levels: [0,30) [30,70) [70,100)But the last value 100 is excluded now. We can include it by setting the include.lowest parameter to TRUE.
cut(x, breaks = c(0, 30, 70, 100),
right = F, include.lowest = T) [1] [0,30) [0,30) [30,70) [70,100] [30,70) [70,100] [70,100] [30,70)
[9] [30,70) [30,70)
Levels: [0,30) [30,70) [70,100]Level names are automatically set. We can rename them with the labels parameter.
cut(x, breaks = c(0, 30, 70, 100),
right = F, include.lowest = T,
labels = c("Low","Medium","High")) [1] Low Low Medium High Medium High High Medium Medium Medium
Levels: Low Medium HighOne of the built-in data sets in R is the mtcars data. We can see its first six lines with the head() function call:
head(mtcars) mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1This is an old data set from 1974, collecting some features of 32 automobiles. For more information, use the command help(mtcars).
The summary() function returns the summary statistics for each numeric field.
summary(mtcars) mpg cyl disp hp
Min. :10.40 Min. :4.000 Min. : 71.1 Min. : 52.0
1st Qu.:15.43 1st Qu.:4.000 1st Qu.:120.8 1st Qu.: 96.5
Median :19.20 Median :6.000 Median :196.3 Median :123.0
Mean :20.09 Mean :6.188 Mean :230.7 Mean :146.7
3rd Qu.:22.80 3rd Qu.:8.000 3rd Qu.:326.0 3rd Qu.:180.0
Max. :33.90 Max. :8.000 Max. :472.0 Max. :335.0
drat wt qsec vs
Min. :2.760 Min. :1.513 Min. :14.50 Min. :0.0000
1st Qu.:3.080 1st Qu.:2.581 1st Qu.:16.89 1st Qu.:0.0000
Median :3.695 Median :3.325 Median :17.71 Median :0.0000
Mean :3.597 Mean :3.217 Mean :17.85 Mean :0.4375
3rd Qu.:3.920 3rd Qu.:3.610 3rd Qu.:18.90 3rd Qu.:1.0000
Max. :4.930 Max. :5.424 Max. :22.90 Max. :1.0000
am gear carb
Min. :0.0000 Min. :3.000 Min. :1.000
1st Qu.:0.0000 1st Qu.:3.000 1st Qu.:2.000
Median :0.0000 Median :4.000 Median :2.000
Mean :0.4062 Mean :3.688 Mean :2.812
3rd Qu.:1.0000 3rd Qu.:4.000 3rd Qu.:4.000
Max. :1.0000 Max. :5.000 Max. :8.000 Even though all data appears as numeric, some columns are actually categorical: "cyl" gives the number of cylinders in the engine, "vs" indicates if the car has a V-shaped engine (0) or a straight (0), "am" indicates if it has automatic transmission (0) or manual (1), "gear" is the number of forward gears, and "carb" is the number of carburetors.
If we convert these columns to factors, summary() and other R functions will give more relevant information about the data.
cyl, gear and carb are ordinal factors (can be ordered meaningfully), so we set ordered=TRUE for them.
mtcars$cyl <- factor(mtcars$cyl, ordered=TRUE)
mtcars$gear <- factor(mtcars$gear, ordered=TRUE)
mtcars$carb <- factor(mtcars$carb, ordered=TRUE)
mtcars$vs <- factor(mtcars$vs)
mtcars$am <- factor(mtcars$am)Now we can use the summary() function to get the counts of categories in each factor field.
summary(mtcars) mpg cyl disp hp drat
Min. :10.40 4:11 Min. : 71.1 Min. : 52.0 Min. :2.760
1st Qu.:15.43 6: 7 1st Qu.:120.8 1st Qu.: 96.5 1st Qu.:3.080
Median :19.20 8:14 Median :196.3 Median :123.0 Median :3.695
Mean :20.09 Mean :230.7 Mean :146.7 Mean :3.597
3rd Qu.:22.80 3rd Qu.:326.0 3rd Qu.:180.0 3rd Qu.:3.920
Max. :33.90 Max. :472.0 Max. :335.0 Max. :4.930
wt qsec vs am gear carb
Min. :1.513 Min. :14.50 0:18 0:19 3:15 1: 7
1st Qu.:2.581 1st Qu.:16.89 1:14 1:13 4:12 2:10
Median :3.325 Median :17.71 5: 5 3: 3
Mean :3.217 Mean :17.85 4:10
3rd Qu.:3.610 3rd Qu.:18.90 6: 1
Max. :5.424 Max. :22.90 8: 1 The "vs" (V-shaped engine or straight) and "am" (Automatic or manual transmission) fields have level values 0 or 1.
levels(mtcars$am)[1] "0" "1"Let’s replace the level values with clearer labels.
levels(mtcars$vs) <- c("V-engine","Standard")
levels(mtcars$am) <- c("Automatic","Manual")summary(mtcars) mpg cyl disp hp drat
Min. :10.40 4:11 Min. : 71.1 Min. : 52.0 Min. :2.760
1st Qu.:15.43 6: 7 1st Qu.:120.8 1st Qu.: 96.5 1st Qu.:3.080
Median :19.20 8:14 Median :196.3 Median :123.0 Median :3.695
Mean :20.09 Mean :230.7 Mean :146.7 Mean :3.597
3rd Qu.:22.80 3rd Qu.:326.0 3rd Qu.:180.0 3rd Qu.:3.920
Max. :33.90 Max. :472.0 Max. :335.0 Max. :4.930
wt qsec vs am gear carb
Min. :1.513 Min. :14.50 V-engine:18 Automatic:19 3:15 1: 7
1st Qu.:2.581 1st Qu.:16.89 Standard:14 Manual :13 4:12 2:10
Median :3.325 Median :17.71 5: 5 3: 3
Mean :3.217 Mean :17.85 4:10
3rd Qu.:3.610 3rd Qu.:18.90 6: 1
Max. :5.424 Max. :22.90 8: 1 When given a factor variable, the plot() function displays a bar plot by default.
plot(mtcars$am)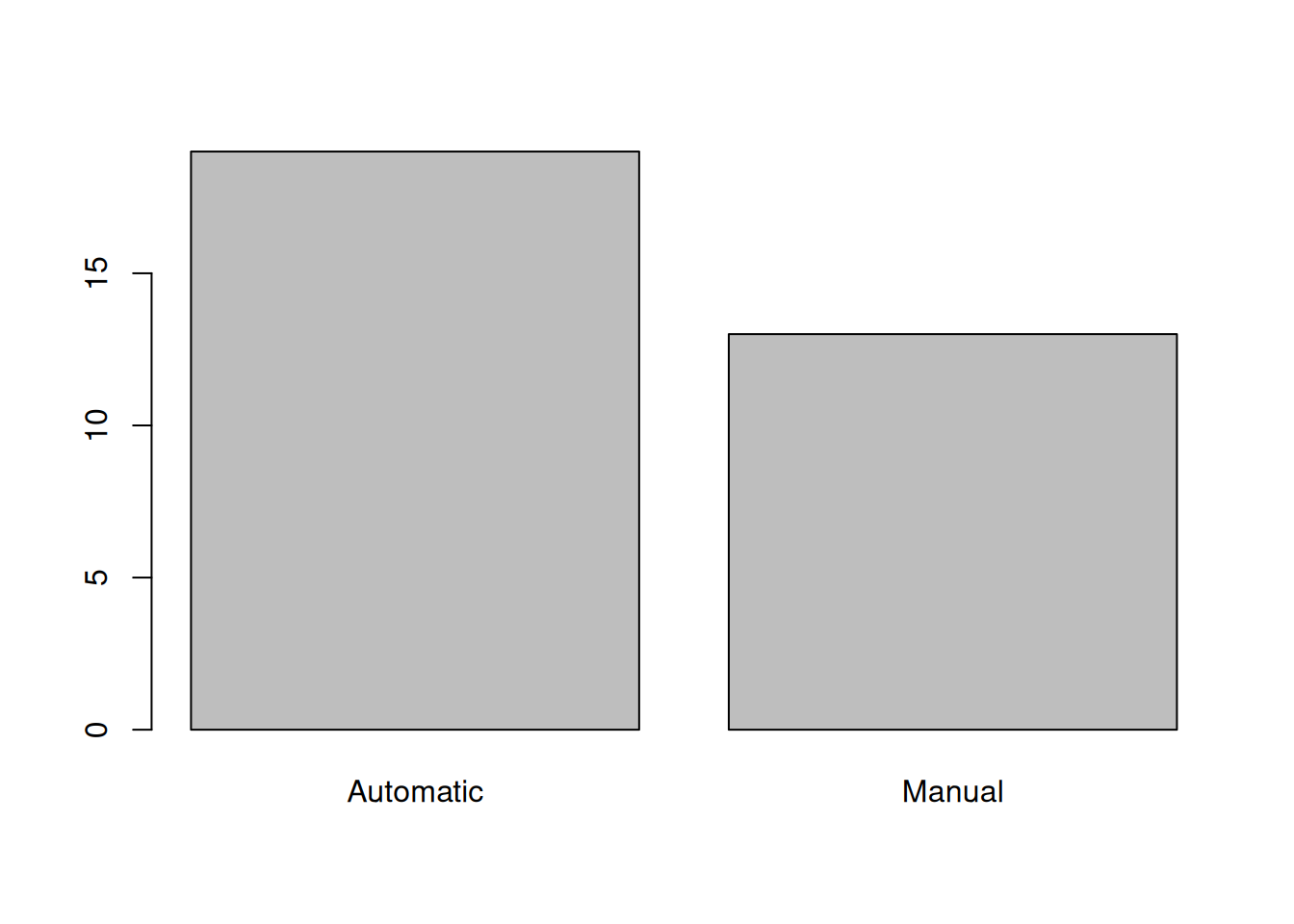
When the x-axis is categorical and the y-axis is numerical, a boxplot is displayed.
plot(x = mtcars$vs, y=mtcars$hp, ylab="Horse power")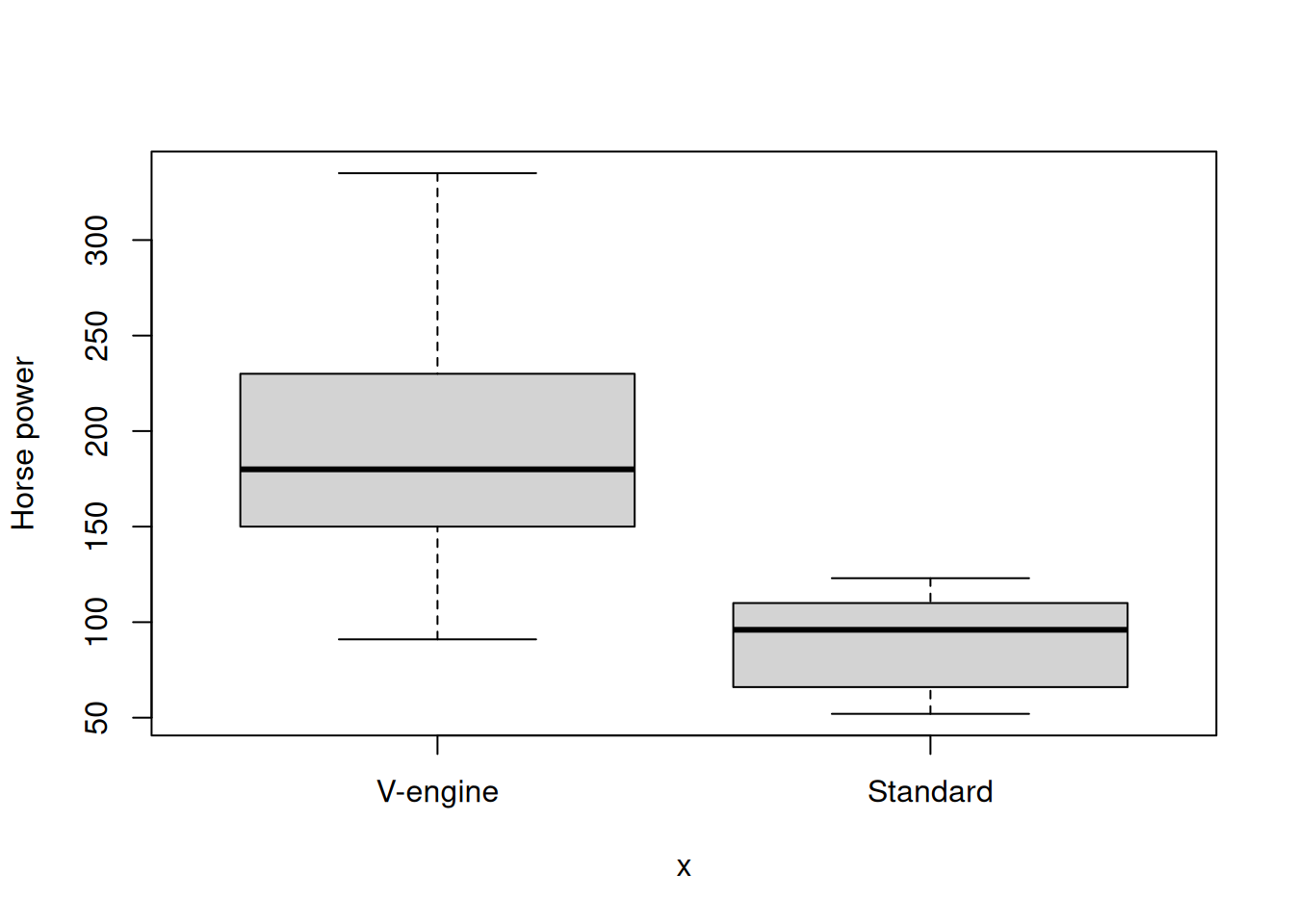
If both axes are categorical, a stacked bar plot is displayed.
plot(x = mtcars$vs, y=mtcars$gear, xlab="Engine type",ylab="Gear")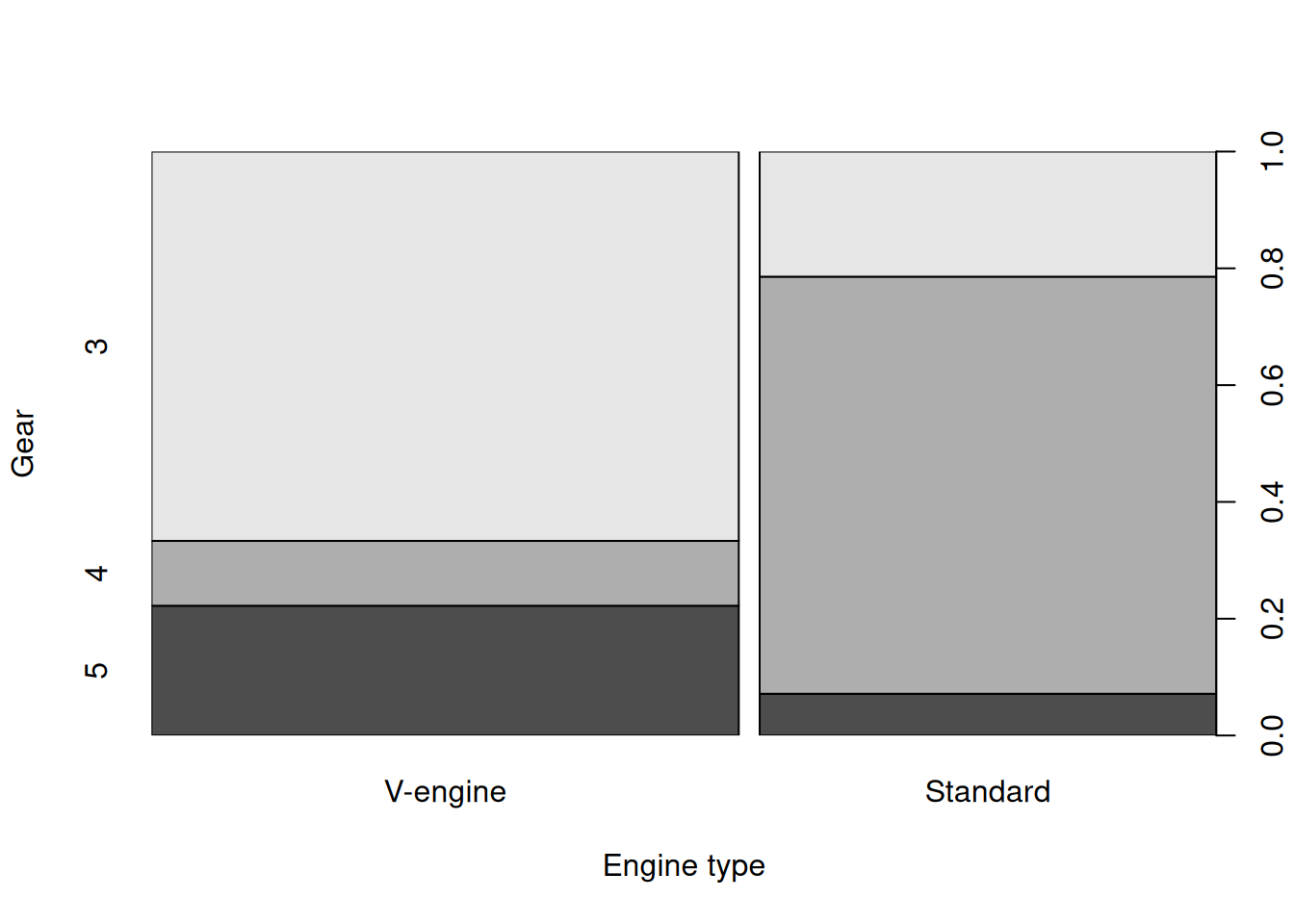
table() functionThe table() function can be used to return counts of elements in each level of a categorical variable.
affils <- c("R","D","D","R","U","D") # political party affiliations
table(affils)affils
D R U
3 2 1 It can be used to create contingency tables, such as two-way tables:
table(mtcars$am, mtcars$vs)
V-engine Standard
Automatic 12 7
Manual 6 7Or three-way tables:
table(mtcars$am, mtcars$vs, mtcars$gear,
dnn=c("Transmission","Engine","Gears")), , Gears = 3
Engine
Transmission V-engine Standard
Automatic 12 3
Manual 0 0
, , Gears = 4
Engine
Transmission V-engine Standard
Automatic 0 4
Manual 2 6
, , Gears = 5
Engine
Transmission V-engine Standard
Automatic 0 0
Manual 4 1tapply() functionThis is more general than table(). Both apply on groups broken by categories, but table() gives only the counts of these groups, while tapply() applies any function to the groups, such as the mean, median, or maximum.
The function call tapply(x, f, func) breaks the vector x by levels given in f and applies the function func on each subgroup.
Given the ages and party affiliations of a group of people, find the average age of people in every party:
ages <- c(25, 26, 55, 37, 21, 42) # ages of some people
affils <- c("R","D","D","R","U","D") # party affiliations of the same people
tapply(ages, affils, mean) D R U
41 31 21 Use the mtcars data again. Get the mean miles-per-gallon for each engine type.
tapply(mtcars$mpg, mtcars$vs, mean)V-engine Standard
16.61667 24.55714 Get the mean miles-per-gallon again, broken by the engine type and transmission type.
tapply(mtcars$mpg, list(mtcars$vs, mtcars$am), mean) Automatic Manual
V-engine 15.05000 19.75000
Standard 20.74286 28.37143Finally, look at the built-in iris database, which stores measurements of sepals and petals of three species of iris flower.
head(iris) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosaLet’s get the mean sepal length of flowers, broken by the species:
tapply(iris$Sepal.Length, iris$Species, mean) setosa versicolor virginica
5.006 5.936 6.588